Method overview
R2DT (which stands for RNA 2D Templates) is a software package that visualises RNA secondary structure in standard layouts representing a wide range of RNAs:
3D-structure based SSU and LSU rRNA from RiboVision
5S and SSU rRNA from CRW
tRNA from GtRNAdb
RNAse P from Ribonuclease P Database
RNA families from Rfam
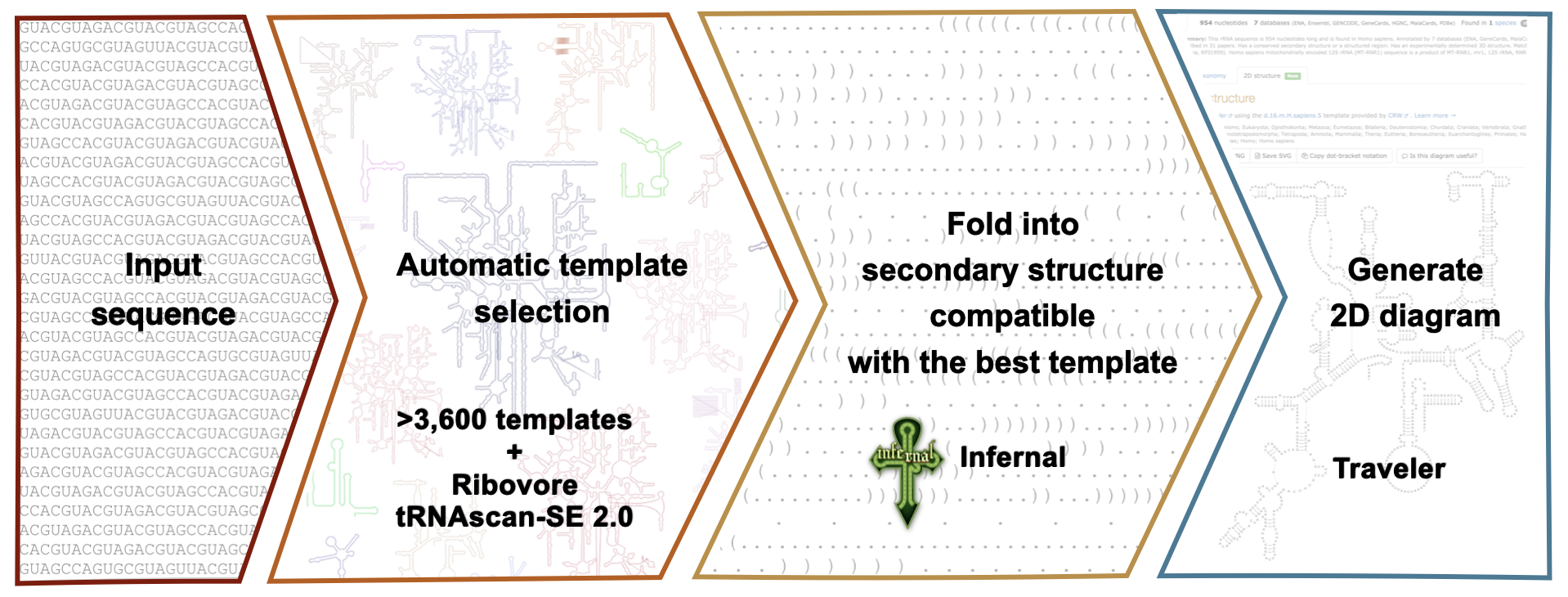
Pipeline
The R2DT pipeline includes the following steps:
Generate a library of covariance models using BPSEQ files from CRW, RiboVision or another source with Infernal. For best results, remove pseudoknots from the secondary structures using RemovePseudoknots from the RNAStructure package.
Select the best matching covariance model for each input sequence using Ribovore or tRNAScan-SE 2.0.
Fold input sequence into a secondary structure compatible with the template using the top scoring covariance model.
Generate secondary structure diagrams using Traveler and the secondary structure layouts.
For a detailed method description see the R2DT paper.