R2DT Version 1.4
We are excited to announce a new release of R2DT. Version 1.4 enables new kinds of visualisations, adds new templates, and brings important technical improvements. Read on to find out more.
Visualising alignment accuracy
R2DT can now visualise additional annotations as data layers on top of the secondary structure diagrams. By default, R2DT displays the expected accuracy of the alignment between the query sequence and the template. The accuracy is calculated by the Infernal software as posterior probabilities.
For example, the following diagram shows the secondary structure of a SAM riboswitch RNA sequence from Bacillus subtilis visualised using the RF00162 Rfam template.
The colored circles highlight the hairpin loop region where Infernal is not 100% confident in the alignment.
Find out how how to visualise your own data, configure colours, and more.
Displaying pseudoknots
Pseudoknot visualisation has been one of the most requested features since R2DT’s launch. Starting with version 1.4, R2DT uses templates derived from Rfam families that have pseudoknots annotated in the consensus secondary structure (SS_cons lines of the Stockholm file format). Upon alignment of a target sequence to the covariance model using Infernal, the pseudoknot annotation is transferred to the alignment using the cmalign program. The following secondary structure of the Cripavirus IRES includes three pseudoknots:
Click to zoom in and see the details.
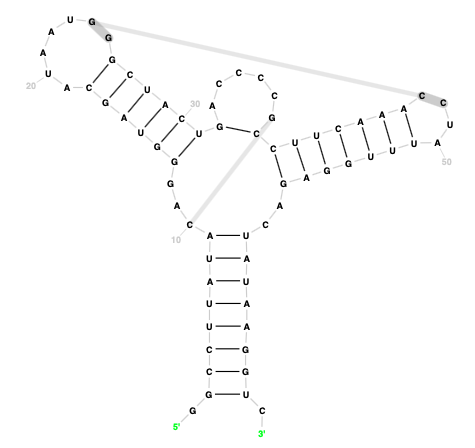
Template-free display
Starting with version 1.4, it is possible to visualise a sequence and its secondary structure without an existing template using a layout generated by R2R. This functionality is useful as a starting point when generating new templates or in cases when the R2DT template library does not yet have a template for a certain RNA. Find more in the docs.
The output files are organised in the standard way and include SVG and RNA 2D JSON Schema files, and can be edited in RNAcanvas and xRNA-React.
New templates
New mito-Vertebrate tRNA and RNAse P templates have been provided by Patricia Chan.
New 3D-based RNAse P template for M. jannaschii and T. maritima submitted by Anton S. Petrov.
The Rfam templates have been updated using Rfam release 14.9.
Other updates
The documentation has been migrated to the Read The Docs platform and can be found at docs.r2dt.bio.
Ribovore, Traveler, Vienna RNA, R-scape, and other dependencies have been updated.
Automated pre-commit software quality checks have been added.
A new process for testing changes using image similarity instead of file identity has been implemented.
R2DT now runs natively on Apple silicon.
The Docker image size has been reduced 5x.
Teamwork makes the dream work
A big thank you to everyone who helped make this release possible, especially:
David Hoksza for adding support for data layers and pseudoknots to the Traveler software,
Eric Nawrocki for help with the Infernal software suite,
Elena Rivas for help with optimising the R-scape installation,
Patricia Chan and the Rfam team for new templates,
Ronny Lorenz for help with Vienna RNA python bindings on Apple silicon,
Anton S. Petrov, Blake Sweeney, and everyone else from the R2DT team.
Last but not least, welcome to a new contributor, Alexey Nayden, who bravely re-engineered the Docker image and developed the new testing setup 👏👏👏
Summary
The new software is now available on GitHub and Docker Hub. The RNAcentral web app and the API will be updated with the new software in the coming weeks. Stay tuned for future releases and feel free to let us know if you have any feedback by raising an issue on GitHub.